DEMO - Working with FLASH data
Last modified by makuadm on 2026-01-07 06:21
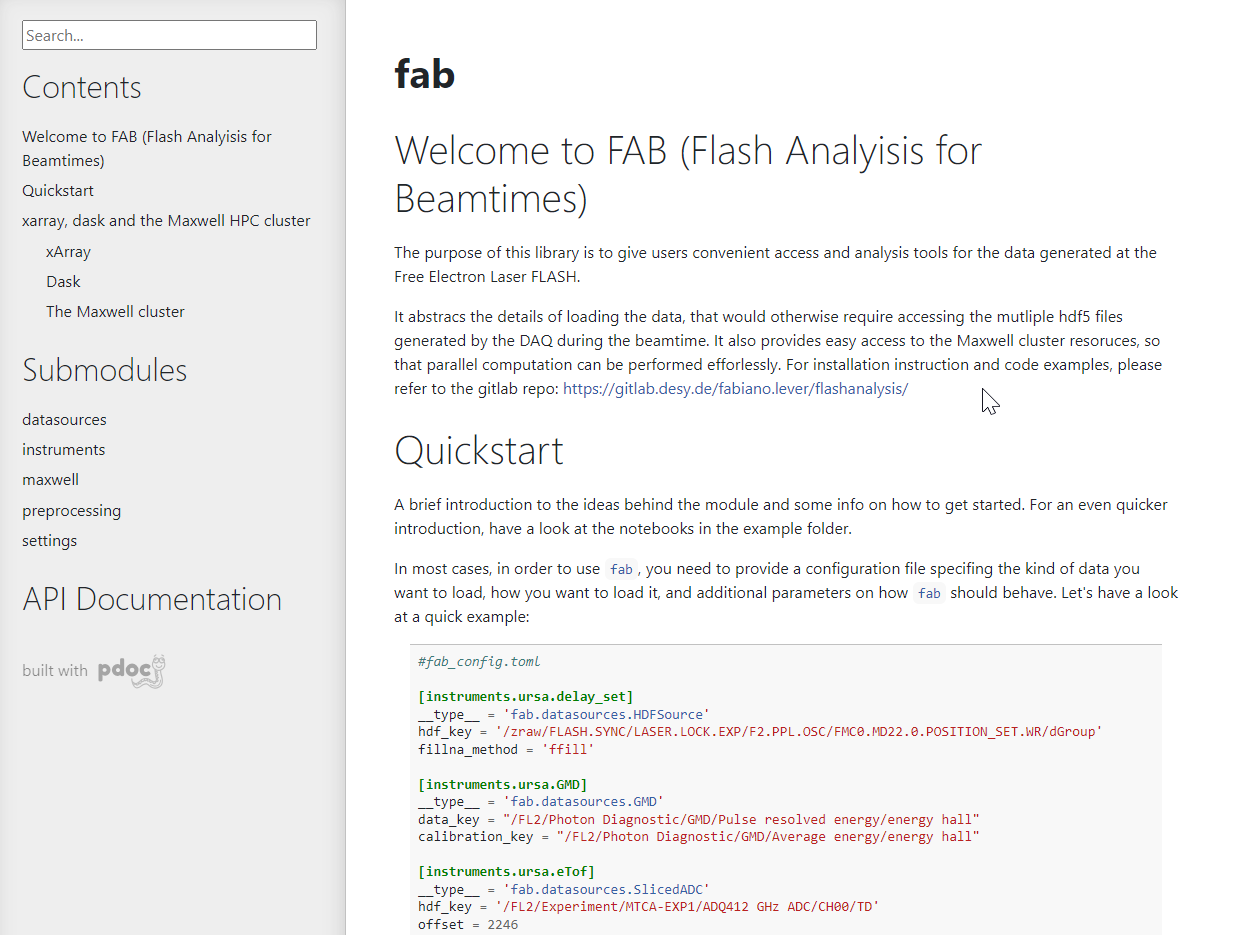
Experimental data is recorded as HDF files[link] on the GPFS file system[link]. The access rights are linked to the user's DESY account and can be managed by the PI via the GAMMA portal[link]. The experimental data can be downloaded via the GAMMA portal, but it is advised to use the DESY computing infrastructure. Access point are via ssh, Maxwell-Display Server[link] or JuyterHub[link]. We recommend using the JupyterHub for data exploration and the SLURM resources for high performances computing - see FAB for easy usage of the infrastructure.